Polysaccharide Storage Myopathy (PSSM)
Summary
Polysaccharide Storage Myopathy (PSSM) is a form of exercise intolerance. The clinical signs manifesting during or after exercise resemble other types of exertional rhabdomyolysis. During an episode, horses are reluctant to move, experience pain, stiffness, and tremors, and sweat profusely. Serum creatine kinase (CK) and aspartate aminotransferase (AST) are elevated, indicating muscle damage. In severe episodes, pigmenturia (coffee-colored urine due to the presence of myoglobin), inability to stand, and death can occur. These symptoms resemble other forms of exercise intolerance such as Recurrent Exertional Rhabdomyolysis (RER), Myofibrillar Myopathy (MFM), and Vacuolar Myopathy (VM). Dietary therapy (elimination of sugars and the addition of protein and fats) and a specific exercise regimen can help to manage symptoms.
Date of Last Update: 08/01/2016
Polysaccharide storage myopathy, type 1
Back to TopSummary
Polysaccharide Storage Myopathy, type 1 (PSSM1) is associated with a semidominant variant of the GYS1 gene that encodes glycogen synthase (GYS1-R309H). The variant form of the enzyme is overactive, causing excess glycogen storage in muscle tissue. Many horses that carry this variant do not show symptoms. Horses affected with both PSSM1 (GYS1-R309H) and Malignant Hyperthermia (MH) (RYR1-R2454G) have higher exercise intolerance than those with PSSM1 alone.
Over 10% of Quarter Horses are affected. Over 60% of Percherons and 90% of Belgian Draft horses are affected. The American Quarter Horse Association requires genetic testing for PSSM1 and four other genetic conditions (GBED, HYPP, MH, and HERDA) when a Quarter Horse is registered.
A substantial fraction of horses that carry the GYS1-R309H variant are asymptomatic. There is some evidence that additional genetic factors modify the development of symptoms in horses carrying the GYS1-R309H variant. EquiSeq is currently searching for variants that affect the development of symptoms in horses carrying the GYS1-R309H variant.
There are cases of PSSM that are not associated with the GYS1-R309H variant. These cases are referred to as PSSM Type 2. The genetic basis of PSSM2 is under investigation. There appear to be multiple subtypes of PSSM2 in different breeds.
In most cases, a definitive diagnosis of PSSM2 requires both a muscle biopsy and a negative DNA test for the GYS1-R309H variant that is associated with PSSM1. This will change as the genetic causes of PSSM2 are identified.
Date of Last Update: 09/06/2016
Results
Understanding the Results
Results of the genetic test for PSSM1 are presented as shown below.
| Polysaccharide Storage Myopathy Type 1 (PSSM1) | ||
|---|---|---|
| n/n | Clear | This horse tested negative for P1 and does not carry the GYS1-R309H mutation identified in Quarter Horses. The horse will not pass on the defect to its offspring. |
| n/P1 | Affected | Both the normal and mutant alleles are present. This horse is positive for the GYS1-R309H (P1) mutation and may develop symptoms of exercise intolerance. |
| P1/P1 | Affected | This horse carries two copies of the GYS1-R309H (P1) mutation and may develop symptoms of exercise intolerance. |
Some horses carrying the GYS1-R309H (P1) mutation never develop symptoms of exercise intolerance, regardless of their diet or exercise program. It is possible that the effect of the GYS1-R309H mutation is modified by other genetic variants.
Disease Name and Genes
PSSM1 is associated with a G to A substitution in the GYS1 gene that causes an Arginine (R) to Histidine (H) substitution at amino acid position 309 in Glycogen Synthase I (GYS1-R309H).
Inheritance
PSSM1 is associated with a semidominant allele of the GYS1 gene (GYS1-R309H) that has incomplete penetrance. The semidominant allele is commonly abbreviated as P1, with the recessive wild-type allele abbreviated as n.

A horse with one copy of the semidominant allele (n/P1) will potentially have symptoms. If this horse is bred to a normal horse (n/n), each foal has a 50% chance of having one copy of the semidominant allele (n/P1) and a 50% chance of having two copies of the normal allele (n/n).

If two horses, each with one copy of the semidominant allele (n/P1), are bred, each foal has a 25% chance of having two copies of the normal allele (n/n), a 50% chance of having one copy of the semidominant allele (n/P1), and a 25% chance of having two copies of the semidominant allele (P1/P1). Because horses with one copy (n/P1) or two copies (P1/P1) of the semidominant allele are affected, each foal will have a 75% chance of potentially being affected.

If a horse with two copies of the semidominant allele (P1/P1) is bred to a normal horse (n/n), all of the foals will have one copy of the semidominant allele (n/P1) and will potentially be affected.
Citations
McCue ME et al. (2008). "Glycogen synthase (GYS1) mutation causes a novel skeletal muscle glycogenosis." Genomics. 91(5):458-66. PMID: 18358695.McCue ME et al. (2008). "Glycogen synthase 1 (GYS1) mutation in diverse breeds with polysaccharide storage myopathy." Journal of Veterinary Internal Medicine. 22(0):1228–1233. PMID: 18691366.
McCue ME et al. (2009). "Polysaccharide storage myopathy phenotype in quarter horse-related breeds is modified by the presence of an RYR1 mutation." Neuromuscular Disorders. 19(0):37–43. PMID: 19056269.
McCue ME et al. (2009). "Comparative skeletal muscle histopathologic and ultrastructural features in two forms of polysaccharide storage myopathy in horses." Vet Pathol. 46(6):1281-1291. PMID: 19605906.
Maile CA et al. (2017). "A highly prevalent equine glycogen storage disease is explained by constitutive activation of a mutant glycogen synthase." Biochim Biophys Acta.. 1861(1):3388-3398. PMID: 27592162.
Polysaccharide storage myopathy, type 2
Back to TopSummary
There are cases of PSSM that are not associated with the GYS1-R309H variant. These cases are referred to as PSSM Type 2. The genetic basis of PSSM2 is under investigation. There appear to be multiple genetic causes of PSSM2 in different breeds.
In most cases, a definitive diagnosis of PSSM2 requires both a muscle biopsy and a negative DNA test for the GYS1-R309H variant (P1) that is associated with PSSM1. This will change as the genetic causes of PSSM2 are identified.
Muscle biopsy of horses with PSSM2 shows abnormal clumping of normally-sized glycogen granules in regions of focal myofibrillar disruption. The primary cause of the defect in PSSM2 therefore appears to be disorganization of myofibrils, rather than a disorder of glycogen metabolism as is the case in PSSM1.
In contrast to PSSM1 and RER, horses with PSSM2 may show levels of serum CK and AST that are at the high end of the normal range. This means that the measurement of serum CK and AST may not be useful as diagnostic criteria for PSSM2.
Researchers at EquiSeq have identified five semidominant genetic variants that cause PSSM2. The results are not yet published in a peer-reviewed academic journal. Prior to publication, the variants has been termed P2, P3, P4, P8, and K1. These are missense alleles of MYOT, FLNC, MYOZ3, PYROXD1, and COL6A3, respectively.
PSSM2 is also associated with two recessive variants of an undisclosed gene designated P5 and P6. P5 and P6 are alleles of the same gene.
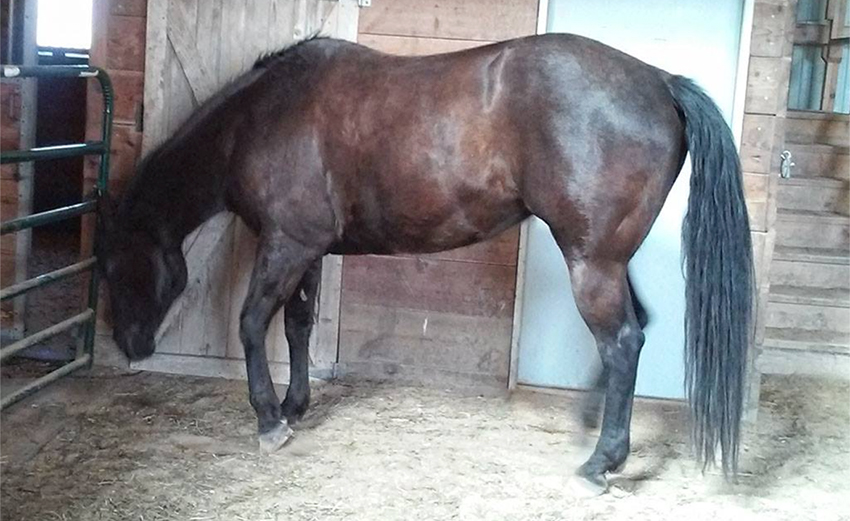

The images above show Gal's Magnificent Blunder (Maggie), a Quarter Horse with PSSM2 born in 1999, in May 2015 (top) and May 2016 (bottom). Maggie is heterozygous for both the P2 (n/P2) and P3 (n/P3) variants and does not carry the P1 variant (n/n). The P3 variant is associated with Myofibrillar Myopathy (MFM). Maggie lost 200 pounds over a period of one year. The muscle wasting is most apparent in the hindquarters.
The video above shows Supernatural, a Thoroughbred heterozygous for both P3 and P4 (n/P3 n/P4). Stiffness in the hindquarters is evident, as is "bunny hopping" and cross-firing in the canter.
Horses that carry two copies of the variants (P2/P2, P3/P3, or P4/P4) are more strongly affected, with an earlier age of onset and more severe symptoms. An early symptom in affected horses is a change in temperament likely related to pain. The horse may suddenly seem reluctant to be saddled or handled in other ways, and resist having a foot raised for the farrier. Another symptom is shifting lameness: the horse appears lame in one leg, which gets better on its own, then lameness appears in another leg. Muscle wasting, especially in the hindquarters (pelvic girdle and proximal limb) and topline (shoulder girdle), may be apparent. The horse may also show localized muscle wasting, divots that look like kick marks. Muscle wasting may produce a rippled or washboard appearance in some areas. Changes in gait are apparent. The most telling sign is at canter, where the horse may "bunny hop" by pulling both rear legs forward at once. In other gaits, the hind limbs may appear stiff with a short gait. Cross-firing (disunited canter) is also seen. "Rope walking," the placement of one foot directly in front of the other as if walking a tightrope, may appear in the rear legs or in all four.
Horses that carry one copy of a variant (n/P2, n/P3, or n/P4) have a predisposition to develop PSSM2. Not every n/P2, n/P3, or n/P4 horse develops symptoms. Those n/P2, n/P3, or n/P4 horses that develop symptoms have a later age of onset (7-10 years) and milder symptoms. The symptoms are the same as those seen in P2/P2, P3/P3, or P4/P4 horses. Development of symptoms appears to be related to whether more than one genetic variant is present in that horse. A horse that is n/P2 n/P3 or n/P2 n/P4 is more likely to develop symptoms than is a horse that is n/P2 but n/n for P3 and P4.
The P8 and K1 genetic variants are expected to interact with P2, P3, and P4 in a similar way. The P5 and P6 genetic variants are expected to interact with other variants in horses homozygous for P5 (P5/P5), homozygous for P6 (P6/P6), or in compound heterozygotes (P5/P6).
Dietary therapy (a diet supplemented with complete protein) and a specific exercise regimen may help to manage the symptoms. This is still an active area of investigation.
Date of Last Update: 10/18/2022
Results
Understanding the Results
Results of the genetic test for the P2 variant of MYOT are presented as shown below. Horses carrying the P2 variant develop symptoms of exercise intolerance as they age. The age of onset and the severity of symptoms is variable.
| Polysaccharide Storage Myopathy Type 2 (PSSM2) | ||
|---|---|---|
| n/n | Clear | This horse tested negative for P2, a PSSM2-predisposing variant. The horse will not pass on the defect to its offspring. |
| n/P2 | Affected | Both the normal and mutant alleles are present. This horse is positive for the P2 variant and may develop symptoms of exercise intolerance. |
| P2/P2 | Affected | This horse carries two copies of the P2 variant. The horse is expected to develop symptoms of exercise intolerance. |
Results of the genetic test for the P3 variant of FLNC are presented as shown below. Horses carrying the P3 variant develop symptoms of exercise intolerance as they age. The age of onset and the severity of symptoms is variable.
| Polysaccharide Storage Myopathy Type 2 (PSSM2) | ||
|---|---|---|
| n/n | Clear | This horse tested negative for P3, a PSSM2-predisposing variant. The horse will not pass on the defect to its offspring. |
| n/P3 | Affected | Both the normal and mutant alleles are present. This horse is positive for the P3 variant and may develop symptoms of exercise intolerance. |
| P3/P3 | Affected | This horse carries two copies of the P3 variant. The horse is expected to develop symptoms of exercise intolerance. |
Results of the genetic test for the P4 variant of MYOZ3 are presented as shown below. Horses carrying the P4 variant develop symptoms of exercise intolerance as they age. The age of onset and the severity of symptoms is variable.
| Polysaccharide Storage Myopathy Type 2 (PSSM2) | ||
|---|---|---|
| n/n | Clear | This horse tested negative for P4, a PSSM2-predisposing variant. The horse will not pass on the defect to its offspring. |
| n/P4 | Affected | Both the normal and mutant alleles are present. This horse is positive for the P4 variant and may develop symptoms of exercise intolerance. |
| P4/P4 | Affected | This horse carries two copies of the P4 variant. The horse is expected to develop symptoms of exercise intolerance. |
Results of the genetic test for the P8 variant of an undisclosed gene are presented as shown below. Horses carrying the P8 variant develop symptoms of exercise intolerance as they age. The age of onset and the severity of symptoms is variable.
| Polysaccharide Storage Myopathy Type 2 (PSSM2) | ||
|---|---|---|
| n/n | Clear | This horse tested negative for P8, a PSSM2-predisposing variant. The horse will not pass on the defect to its offspring. |
| n/P8 | Affected | Both the normal and mutant alleles are present. This horse is positive for the P8 variant and may develop symptoms of exercise intolerance. |
| P8/P8 | Affected | This horse carries two copies of the P8 variant. The horse is expected to develop symptoms of exercise intolerance. |
Results of the genetic test for the K1 variant of an undisclosed gene are presented as shown below. Horses carrying the K1 variant develop symptoms of exercise intolerance as they age. The age of onset and the severity of symptoms is variable.
| Polysaccharide Storage Myopathy Type 2 (PSSM2) | ||
|---|---|---|
| n/n | Clear | This horse tested negative for K1, a PSSM2-predisposing variant. The horse will not pass on the defect to its offspring. |
| n/K1 | Affected | Both the normal and mutant alleles are present. This horse is positive for the K1 variant and may develop symptoms of exercise intolerance. |
| K1/K1 | Affected | This horse carries two copies of the K1 variant. The horse is expected to develop symptoms of exercise intolerance. |
Results of the genetic tests for the P5 and P6 variants of an undisclosed gene are presented as shown below. Horses carrying the P5 or P6 variants develop symptoms of exercise intolerance as they age. The age of onset and the severity of symptoms is variable.
| Polysaccharide Storage Myopathy Type 2 (PSSM2) | ||
|---|---|---|
| N/N | Clear | This horse tested negative for P5, a PSSM2-predisposing variant. The horse will not pass on the defect to its offspring. |
| N/P5 | Carrier | Both the normal and mutant alleles are present. This horse is positive for the P5 variant and is not expected to develop symptoms of exercise intolerance but may pass the defect on to its offspring. |
| N/P6 | Carrier | Both the normal and mutant alleles are present. This horse is positive for the P6 variant and is not expected to develop symptoms of exercise intolerance but may pass the defect on to its offspring. |
| P5/P5 | Affected | This horse carries two copies of the P5 variant. The horse is expected to develop symptoms of exercise intolerance. |
| P6/P6 | Affected | This horse carries two copies of the P6 variant. The horse is expected to develop symptoms of exercise intolerance. |
| P5/P6 | Affected | This horse carries is a compound heterozygote for the P5 and P6 variants. The horse is expected to develop symptoms of exercise intolerance. |
What You Can Do
Horses that test positive for P2, P3, P4, P8, or K1 should receive dietary supplementation with complete protein (whey), complementary protein (soy) or with specific amino acids that are typically limiting in plant protein (lysine, methionine, and threonine). This is a management strategy and not a treatment or cure. This management strategy may also be useful for P5/P5, P6/P6, and P5/P6 horses.
Disease Name and Genes
PSSM2 is associated with semidominant variants (P2, P3, P4, P8, and K1) of MYOT, FLNC, MYOZ3, PYROXD1, and COL6A3, respectively. PSSM2 may also be associated with recessive variants of an undisclosed gene designated P5 and P6.
| Horse Genes Associated with PSSM2 and Human Diseases Associated with Variants of the Orthologous Genes | Variant | Inheritance | Gene Symbol | Gene Name | Variant | Associated Human Disease |
|---|---|---|---|---|---|
| P2 | Semidominant | MYOT | Myotilin | MYOT-S232P chr14:37,818,823 A/G in EquCab3.0 | Myofibrillar Myopathy 3 Limb-Girdle Muscular Dystrophy 1A |
| P3 | Semidominant | FLNC | Filamin C | FLNC-E753K chr4:83,837,774 G/A in EquCab3.0 FLNC-A1207T chr4:83,840,299 G/A in EquCab3.0 | Myofibrillar Myopathy 5 Distal Myopathy 4 |
| P4 | Semidominant | MYOZ3 | Myozenin 3 | MYOZ3-S42L chr14:26,710,261 G/A in EquCab3.0 | none |
| P8 | Semidominant | PYROXD1 | Pyridine nucleotide-disulfide oxidoreductase domain 1 | PYROXD1-D492H chr6:48,924,749 G/C in EquCab3.0 | Myofibrillar Myopathy 8 |
| K1 | Semidominant | COL6A3 | Collagen alpha-3(VI) chain | COL6A3-G2178A chr6:23,416,882 C/G in EquCab3.0 | Bethlem Myopathy Ullrich Congenital Muscular Dystrophy (LGMDR22) |
| P5, P6 | Recessive | undisclosed | undisclosed | undisclosed | undisclosed |
Inheritance
PSSM2 is associated with semidominant variants (P2, P3, P4, P8 and K1) of MYOT, FLNC, MYOZ3, PYROXD1, and COL6A3, respectively. The semidominant alleles are abbreviated as P2, P3, P4, P8, and K1 with the recessive wild-type alleles abbreviated as n.
The illustrations below show the inheritance of P2 as an example. P3, P4, P8, and K1 are inherited in the same way. The five variants are inherited independently, and a horse may have more than one, as shown in the photographs above, of a horse that has the genotype n/P2 n/P3.

A horse with one copy of the semidominant allele (n/P2) will potentially have symptoms. If this horse is bred to a normal horse (n/n), each foal has a 50% chance of having one copy of the semidominant allele (n/P2) and a 50% chance of having two copies of the normal allele (n/n).

If two horses, each with one copy of the semidominant allele (n/P2), are bred, each foal has a 25% chance of having two copies of the normal allele (n/n), a 50% chance of having one copy of the semidominant allele (n/P2), and a 25% chance of having two copies of the semidominant allele (P2/P2). Because horses with one copy (n/P2) or two copies (P2/P2) of the semidominant allele are affected, each foal will have a 75% chance of potentially being affected.

If a horse with two copies of the semidominant allele (P2/P2) is bred to a normal horse (n/n), all of the foals will have one copy of the semidominant allele (n/P2) and will potentially be affected.
PSSM2 is also caused by a recessive mutation in an undisclosed gene. The recessive allele is abbreviated as P5, with the dominant wild-type allele abbreviated as N.

Carriers of the recessive allele (N/P5) have no symptoms of the disease. If two such carriers are bred, each foal has a 25% chance of having two copies of the normal allele (N/N), a 50% chance of being a carrier (N/P5), and a 25% chance of being affected (P5/P5).

If a carrier of the recessive allele (N/P5) is bred to a normal horse (N/N), each foal has a 50% chance of having two copies of the normal allele (N/N) and a 50% chance of being a carrier (N/P5).
Citations
McCue ME et al. (2009). "Comparative skeletal muscle histopathologic and ultrastructural features in two forms of polysaccharide storage myopathy in horses." Vet Pathol. 46(6):1281-1291. PMID: 19605906.Valberg SJ et al. (2016). "Suspected myofibrillar myopathy in Arabian horses with a history of exertional rhabdomyolysis." Equine Vet J.. 48(5):548-556. PMID: 26234161.
Lewis SS et al. (2017). "Clinical characteristics and muscle glycogen concentrations in warmblood horses with polysaccharide storage myopathy" Am J Vet Res. 78(11):1305-1312. PMID: 29076373.
Williams ZJ et al. (2018). "Muscle glycogen concentrations and response to diet and exercise regimes in Warmblood horses with type 2 Polysaccharide Storage Myopathy" PLOS One. 13(9):e0203467. PMID:30183782.





